23andMe Investor Presentation Deck
The Vast Majority of GWAS Discoveries Can
be Made Without Large-scale Sequencing
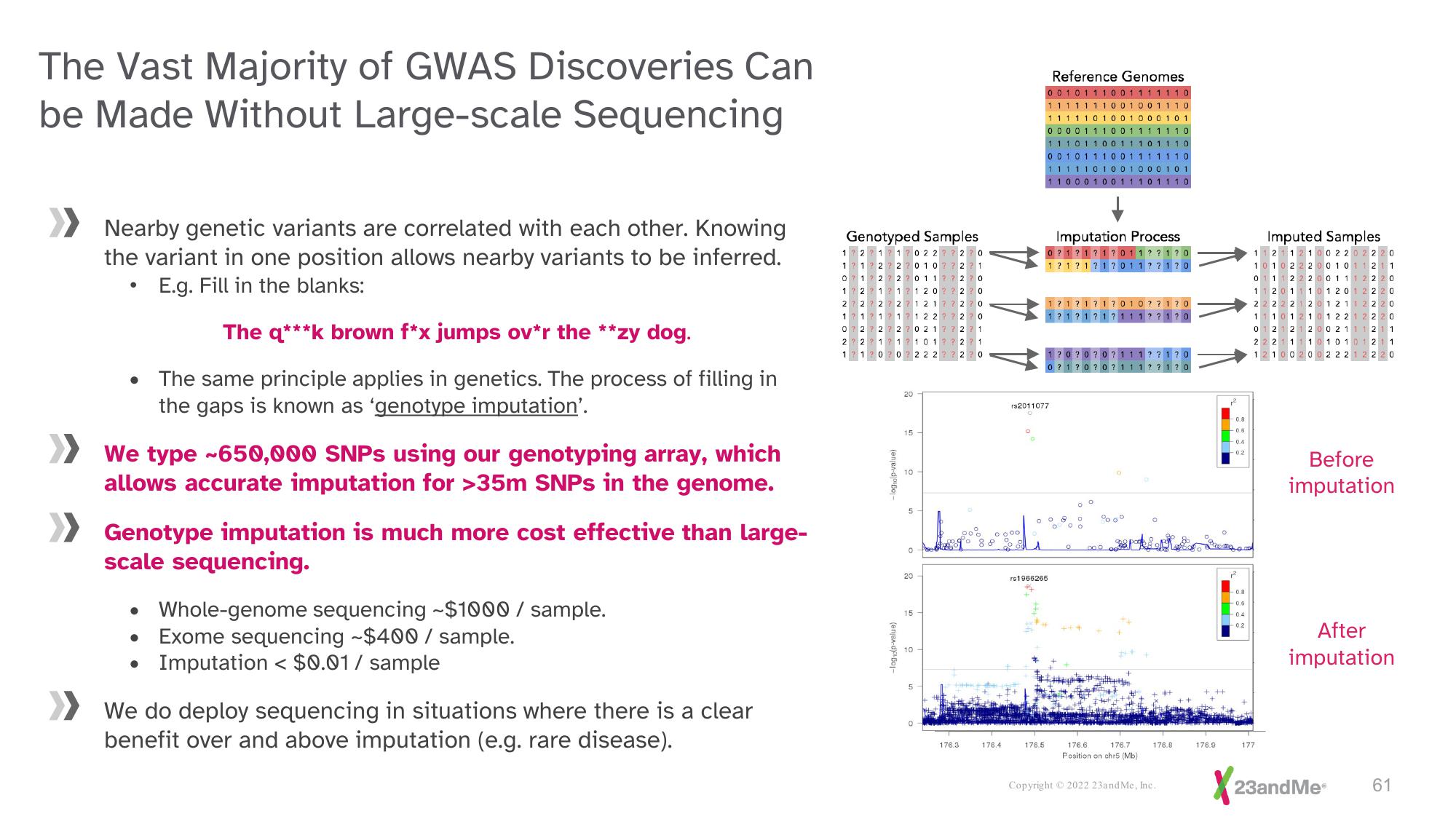
Nearby genetic variants are correlated with each other. Knowing
the variant in one position allows nearby variants to be inferred.
E.g. Fill in the blanks:
The q***k brown f*x jumps ov*r the **zy dog.
The same principle applies in genetics. The process of filling in
the gaps is known as 'genotype imputation'.
We type ~650,000 SNPs using our genotyping array, which
allows accurate imputation for >35m SNPs in the genome.
Genotype imputation is much more cost effective than large-
scale sequencing.
• Whole-genome sequencing -$1000/sample.
Exome sequencing ~$400 / sample.
Imputation < $0.01/ sample
We do deploy sequencing in situations where there is a clear
benefit over and above imputation (e.g. rare disease).
Genotyped Samples
21
022?? 2 0
1 12 22 2 010 2
071227 22 011?? 2 0
17221212 120 2 0
27272727121 270
121212121 222 22 0
1
07272727021772
21 1 101
2
1 1 0 0 222
-logiolp-value)
-log/p-value)
20
15-
o
5
0
20-
15
5
0
2 1
2
0
176.3
go
176.4
0010111001111110
1 1 1 1 1 1 1 0 0 1 0 0 1 1 1 0
1 1 1 1 1 0 1 0 0 1 0 0 0 1 0 1
0 0 0 0 1 1 1 0 0 1 1 1 1 1 10
1110110 011101 110
0010111001111110
1111101001000101
1 1 0 0 0 1 0 0 1 1 1 0 1 1 1 0
rs2011077
rs1966265
Reference Genomes
Imputation Process
0?1? 11
1701
?1? 0
1 2 1 2 1 2 1 2011 ? ? 120
176.5
0 0.00
88 A
1 2 1 2 1 2 1 7 0 1 0 ? ? 1? 0
1?1?1?1? 111 ??1? 0
1202020?111 ? ? 120
0?1?0?0?111? ? 120
008 0
0 0 0
0
%0°
176.6.
176.7
Position on chr5 (Mb)
176.8
Copyright © 2022 23and Me, Inc.
176.9
Imputed Samples
1
2
1 10022
1 102220010
2 0
2 1
0 112220011 2 0
11201110120122 0
2222212012112220
1110121012212220
0121212002111211
2221111 101
1210020 222
177
2 1
2 0
Before
imputation
After
imputation
23andMe
61View entire presentation